
The protein encoded by this gene acts as a homotetramer to catalyze the conversion of homocysteine to cystathionine, the first step in the transsulfuration pathway. The encoded protein is allosterically activated by adenosyl-methionine and uses pyridoxal phosphate as a cofactor. Defects in this gene can cause cystathionine beta-synthase deficiency (CBSD), which can lead to homocystinuria. This gene is a major contributor to cellular hydrogen sulfide production. Multiple alternatively spliced transcript variants have been found for this gene. [provided by RefSeq, Feb 2016]
- Cysteine biosynthetic process from serine
- L-serine metabolic process
- L-serine catabolic process
- Metabolic process
- Cellular amino acid biosynthetic process
- Homocystinuria due to cystathionine beta-synthase deficiency
- Homocystinuria
- Homocysteinemia
- Hypermethioninemia
- Pyridoxine deficiency anemia
- General Aortopathy
Based on Ayass Bioscience, LLC Data Analysis
CBS Localizations – Subcellular Localization Database
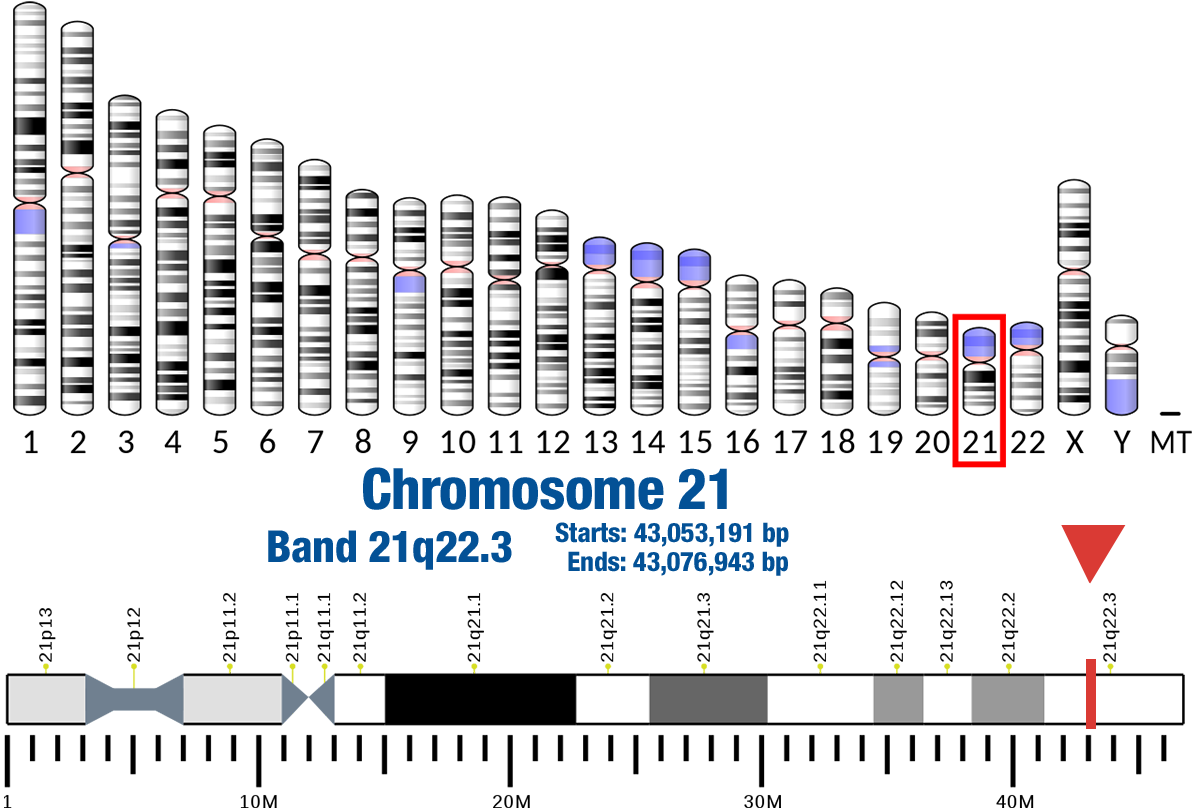
Gene Location
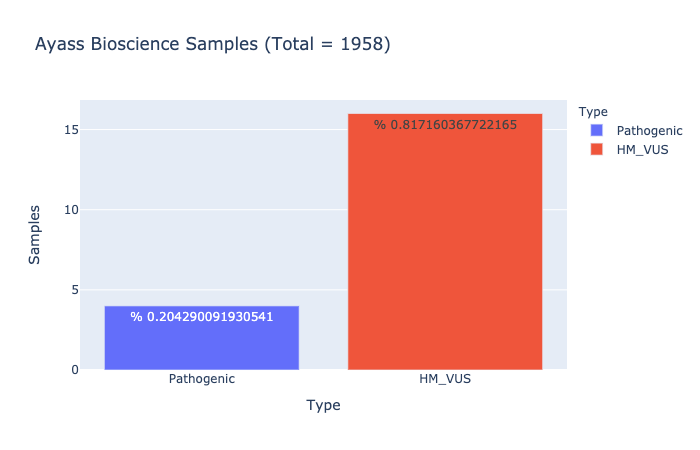
Pathogenic Prevalence
% 0.204290091930541
Ratio of samples with at least 1 pathogenic variant (Computed from Ayass Bioscience Samples)
HM-VUS Prevalence
% 0.817160367722165
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
Pathogenic Variants
G=0.00783(730/93287,ALFAProject)
G=0.00025(20/78612,PAGE_STUDY)
G=0.00083(26/31224,GnomAD)
G=0.0002(1/5008,1000G)
G=0.0034(10/2922,KOREAN)
G=0.011(11/998,GoNL)
G=0.000(0/304,HapMap)
G=0.069(15/216,Qatari)
A=0.50(28/56,SGDP_PRJ)
G=0.50(28/56,SGDP_PRJ)
High/Med VUS Variants
T=0.000155(39/251400,GnomAD_exome)
T=0.000157(19/121292,ExAC)
T=0.00026(24/92796,ALFAProject)
T=0.00001(1/78694,PAGE_STUDY)
T=0.00025(8/31396,GnomAD)
T=0.0002(1/4480,Estonian)
T=0.000008(2/247926,GnomAD_exome)
T=0.000025(3/118276,ExAC)
T=0.00008(1/13006,GO-ESP)
T=0.0000(0/6394,ALFAProject)
G=0.00783(730/93287,ALFAProject)
G=0.00025(20/78612,PAGE_STUDY)
G=0.00083(26/31224,GnomAD)
G=0.0002(1/5008,1000G)
G=0.0034(10/2922,KOREAN)
G=0.011(11/998,GoNL)
G=0.000(0/304,HapMap)
G=0.069(15/216,Qatari)
A=0.50(28/56,SGDP_PRJ)
G=0.50(28/56,SGDP_PRJ)
T=0.000334(56/167900,GnomAD_exome)
T=0.00029(27/92640,ALFAProject)
T=0.00033(26/78610,PAGE_STUDY)
T=0.00026(8/31360,GnomAD)
T=0.00030(7/23348,ExAC)
T=0.0004(2/5008,1000G)
T=0.0000(0/3854,ALSPAC)
T=0.0005(2/3708,TWINSUK)
T=0.002(1/534,MGP)

