
This gene product is the main apolipoprotein of chylomicrons and low density lipoproteins (LDL), and is the ligand for the LDL receptor. It occurs in plasma as two main isoforms, apoB-48 and apoB-100: the former is synthesized exclusively in the gut and the latter in the liver. The intestinal and the hepatic forms of apoB are encoded by a single gene from a single, very long mRNA. The two isoforms share a common N-terminal sequence. The shorter apoB-48 protein is produced after RNA editing of the apoB-100 transcript at residue 2180 (CAA->UAA), resulting in the creation of a stop codon, and early translation termination. Mutations in this gene or its regulatory region cause hypobetalipoproteinemia, normotriglyceridemic hypobetalipoproteinemia, and hypercholesterolemia due to ligand-defective apoB, diseases affecting plasma cholesterol and apoB levels. [provided by RefSeq, Dec 2019]
- Retinoid metabolic process
- In utero embryonic development
- Toll-like receptor signaling pathway
- Lipid metabolic process
- Triglyceride mobilization
- Hypobetalipoproteinemia, familial, 1
- Hypercholesterolemia, familial, 2
- Abetalipoproteinemia
- Homozygous familial hypercholesterolemia
- Hypercholesterolemia, familial, 1
- Familial Hypercholesterolemia
Based on Ayass Bioscience, LLC Data Analysis
APOB Localizations – Subcellular Localization Database
Gene Location

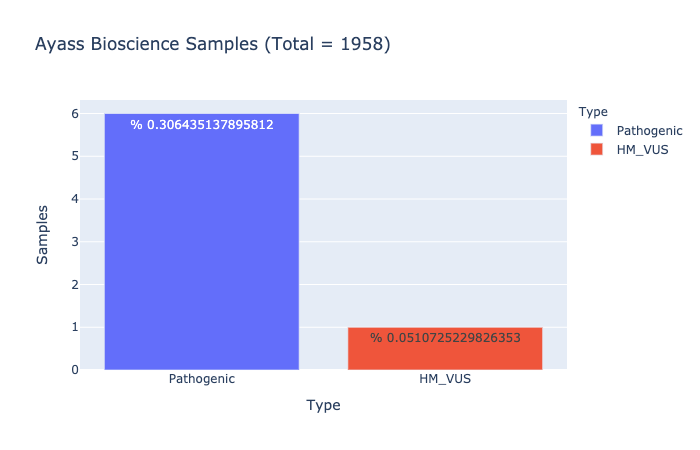
Pathogenic Prevalence
% 0.306435137895812
Ratio of samples with at least 1 pathogenic variant (Computed from Ayass Bioscience Samples)
HM-VUS Prevalence
% 0.0510725229826353
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
Pathogenic Variants
A=0.000012(3/251042,GnomAD_exome)
A=0.000016(2/125568,TOPMED)
A=0.000017(2/121004,ExAC)
A=0.00015(2/13006,GO-ESP)
A=0.0000(0/8586,ALFAProject)
A=0.0000(0/3854,ALSPAC)
A=0.0003(1/3708,TWINSUK)
delCTG=0.000384(96/250322,GnomAD_exome)
delCTG=0.000191(24/125568,TOPMED)
delCTG=0.000512(62/121024,ExAC)
delCTG=0.00025(8/31394,GnomAD)
delCTG=0.00040(5/12518,GO-ESP)
delCTG=0.00018(2/11176,ALFAProject)
delCTG=0.0007(3/4480,Estonian)
delCTG=0.0005(2/3854,ALSPAC)
delCTG=0.0005(2/3708,TWINSUK)
delCTG=0.001(1/998,GoNL)
delAAAGA=0.000004(1/250480,GnomAD_exome)
delAAAGA=0.00032(4/12516,GO-ESP)
T=0.000275(69/250764,GnomAD_exome)
T=0.001338(168/125568,TOPMED)
T=0.000231(28/121182,ExAC)
T=0.00042(40/94988,ALFAProject)
T=0.00014(11/78694,PAGE_STUDY)
T=0.00045(14/31398,GnomAD)
T=0.0013(6/4480,Estonian)
T=0.0003(1/3854,ALSPAC)
T=0.0003(1/3708,TWINSUK)
T=0.0003(1/2922,KOREAN)
T=0.000(0/324,HapMap)

