
This gene encodes lysosomal alpha-glucosidase, which is essential for the degradation of glycogen to glucose in lysosomes. The encoded preproprotein is proteolytically processed to generate multiple intermediate forms and the mature form of the enzyme. Defects in this gene are the cause of glycogen storage disease II, also known as Pompe’s disease, which is an autosomal recessive disorder with a broad clinical spectrum. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jan 2016]
- Maltose metabolic process
- Regulation of the force of heart contraction
- Diaphragm contraction
- Heart morphogenesis
- Carbohydrate metabolic process
- Glycogen storage disease ii
- Glycogen storage disease
- Glycogen storage disease due to acid maltase deficiency, infantile onset
- Glycogen storage disease due to acid maltase deficiency, late-onset
- Isolated elevated serum creatine phosphokinase levels
- Hypertrophic Cardiomyopathy
- General Cardiomyopathy
Based on Ayass Bioscience, LLC Data Analysis
GAA Localizations – Subcellular Localization Database
Gene Location
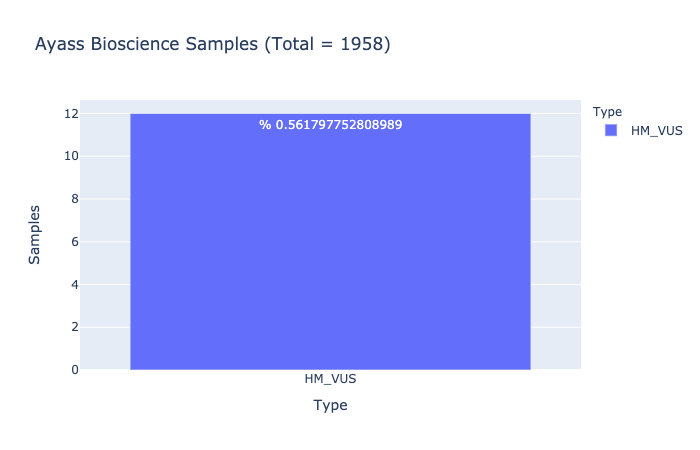
HM-VUS Prevalence
% 0.561797752808989
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
High/Med VUS Variants
T=0.000206(51/247128,GnomAD_exome)
T=0.000287(36/125568,TOPMED)
T=0.000209(25/119472,ExAC)
T=0.00034(26/75456,ALFAProject)
T=0.00020(6/30738,GnomAD)
T=0.00038(5/13006,GO-ESP)
T=0.0004(2/5008,1000G)
T=0.0002(1/4478,Estonian)
T=0.002(1/600,NorthernSweden)
C=0.000390(49/125568,TOPMED)
C=0.00023(18/78700,PAGE_STUDY)
C=0.00031(4/13006,GO-ESP)
C=0.00018(2/11174,ALFAProject)
C=0.0004(2/5008,1000G)
C=0.0004(2/4480,Estonian)
C=0.0003(1/3854,ALSPAC)
C=0.0008(3/3708,TWINSUK)
C=0.0007(2/2918,KOREAN)
C=0.0005(1/1832,Korea1K)
C=0.001(1/998,GoNL)
C=0.005(3/600,NorthernSweden)
C=0.002(1/534,MGP)
delT=0.000096(23/239740,GnomAD_exome)
delT=0.000159(20/125568,TOPMED)
delT=0.000070(8/113628,ExAC)
delT=0.00013(4/31380,GnomAD)
delT=0.00016(2/12176,GO-ESP)
delT=0.00027(3/11176,ALFAProject)
G=0.000012(3/242552,GnomAD_exome)
G=0.000008(1/125568,TOPMED)
G=0.00001(1/88008,ExAC)
G=0.00003(1/31364,GnomAD)
G=0.00018(2/11176,ALFAProject)
T=0.000175(43/246224,GnomAD_exome)
T=0.000510(64/125568,TOPMED)
T=0.000215(25/116512,ExAC)
T=0.00098(77/78180,PAGE_STUDY)
T=0.00004(3/77536,ALFAProject)
T=0.00048(15/31342,GnomAD)
T=0.0010(5/5008,1000G)
A=0.000032(8/248242,GnomAD_exome)
A=0.000024(3/125568,TOPMED)
A=0.000036(4/111856,ExAC)
A=0.00015(2/13000,GO-ESP)
A=0.0000(0/3870,ALFAProject)
delC=0.000029(7/244672,GnomAD_exome)
delC=0.000032(4/125568,TOPMED)
delC=0.000037(4/106676,ExAC)
delC=0.00006(2/31362,GnomAD)
delC=0.00009(1/11176,ALFAProject)

