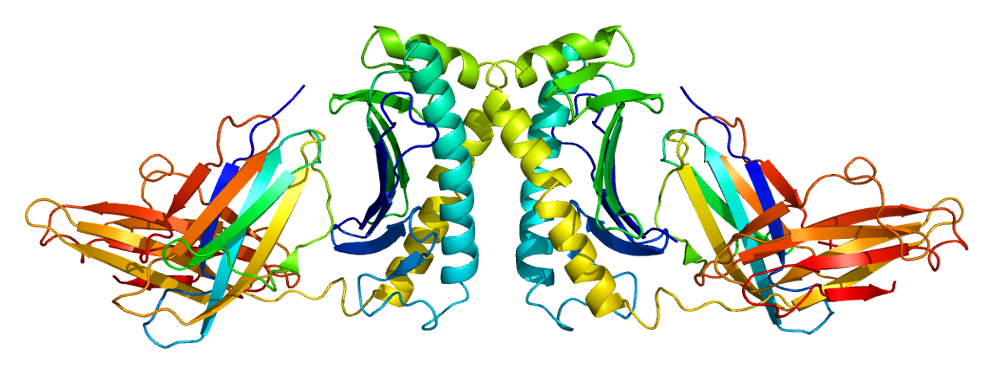
The protein encoded by this gene is a membrane protein that is similar to MHC class I-type proteins and associates with beta2-microglobulin (beta2M). It is thought that this protein functions to regulate iron absorption by regulating the interaction of the transferrin receptor with transferrin. The iron storage disorder, hereditary haemochromatosis, is a recessive genetic disorder that results from defects in this gene. At least nine alternatively spliced variants have been described for this gene. Additional variants have been found but their full-length nature has not been determined. [provided by RefSeq, Jul 2008]
- Positive regulation of T cell mediated cytotoxicity
- NOT antigen processing and presentation of peptide antigen via MHC class I
- Antigen processing and presentation of endogenous peptide antigen via MHC class Ib
- Negative regulation of T cell antigen processing and presentation
- Negative regulation of T cell cytokine production
- Hemochromatosis, type 1
- Transferrin serum level quantitative trait locus 2
- Microvascular complications of diabetes 7
- Variegate porphyria
- Porphyria cutanea tarda
- General Cardiomyopathy
- Dilated Cardiomyopathy
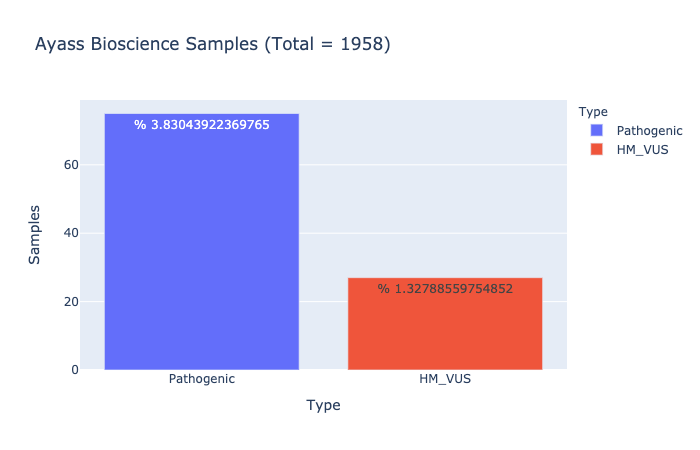
Based on Ayass Bioscience, LLC Data Analysis
HFE Localizations – Subcellular Localization Database
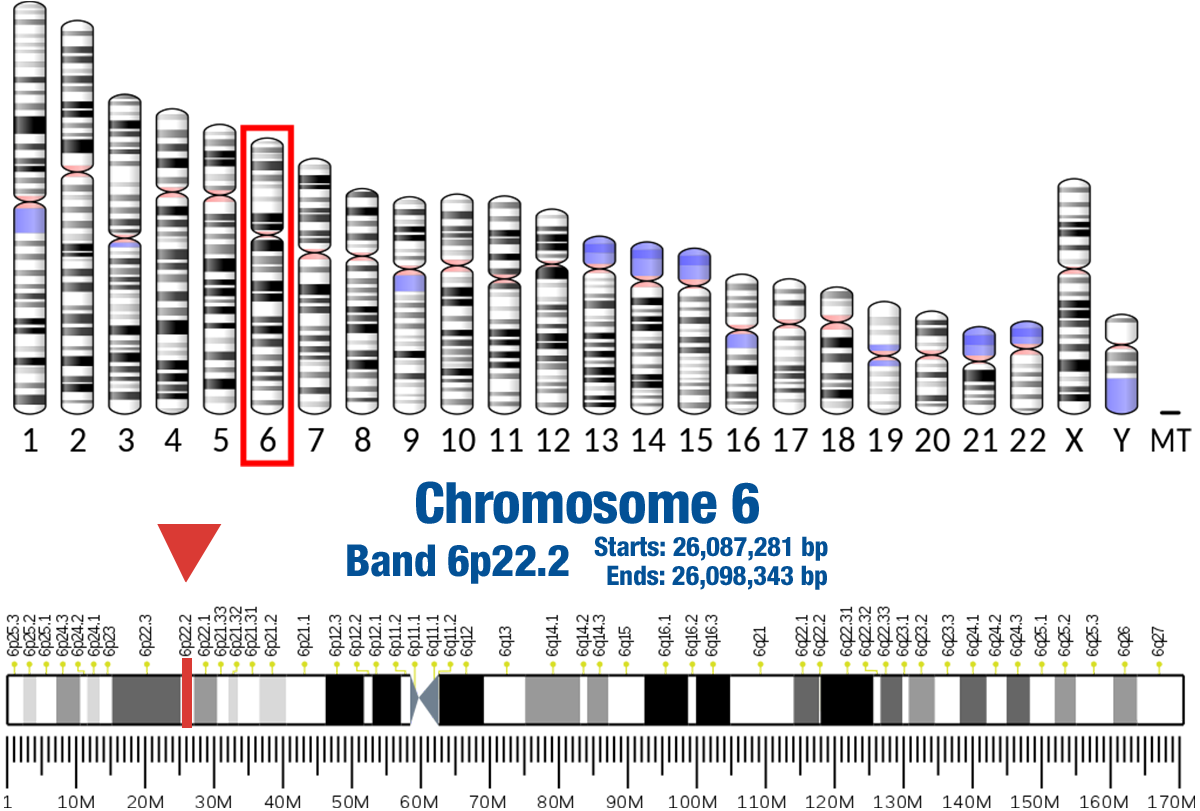
Gene Location
Pathogenic Prevalence
% 3.83043922369765
Ratio of samples with at least 1 pathogenic variant (Computed from Ayass Bioscience Samples)
HM-VUS Prevalence
% 1.32788559754852
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
Pathogenic Variants
G=0.109240(27472/251484,GnomAD_exome)
G=0.101156(12702/125568,TOPMED)
G=0.106599(12942/121408,ExAC)
G=0.06797(5349/78702,PAGE_STUDY)
G=0.09949(3120/31360,GnomAD)
G=0.13586(1977/14552,ALFAProject)
G=0.0731(366/5008,1000G)
G=0.1373(615/4480,Estonian)
G=0.1365(526/3854,ALSPAC)
G=0.1429(530/3708,TWINSUK)
G=0.0488(143/2930,KOREAN)
G=0.0448(82/1832,Korea1K)
G=0.135(135/998,GoNL)
G=0.033(20/614,Vietnamese)
G=0.138(83/600,NorthernSweden)
G=0.199(106/534,MGP)
G=0.102(31/304,FINRISK)
G=0.076(23/302,HapMap)
G=0.130(28/216,Qatari)
C=0.49(43/88,SGDP_PRJ)
G=0.15(6/40,GENOME_DK)
C=0.50(7/14,Siberian)
G=0.50(7/14,Siberian)
A=0.033212(8344/251236,GnomAD_exome)
A=0.053369(7701/144298,ALFAProject)
A=0.038258(4804/125568,TOPMED)
A=0.032432(3937/121394,ExAC)
A=0.01389(1093/78702,PAGE_STUDY)
A=0.03825(1200/31372,GnomAD)
A=0.0126(63/5008,1000G)
A=0.0426(191/4480,Estonian)
A=0.0794(306/3854,ALSPAC)
A=0.0690(256/3708,TWINSUK)
A=0.0072(15/2084,HGDP_Stanford)
A=0.0189(23/1214,HapMap)
A=0.0201(22/1094,Daghestan)
A=0.054(54/998,GoNL)
A=0.018(11/626,Chileans)
A=0.075(45/600,NorthernSweden)
A=0.032(17/534,MGP)
A=0.033(10/304,FINRISK)
A=0.005(1/216,Qatari)
A=0.03(1/40,GENOME_DK)
G=0.50(6/12,SGDP_PRJ)
A=0.50(6/12,SGDP_PRJ)
G=0.5(1/2,Siberian)
A=0.5(1/2,Siberian)
T=0.010239(2575/251490,GnomAD_exome)
T=0.009573(1202/125568,TOPMED)
T=0.010090(1225/121410,ExAC)
T=0.00410(323/78702,PAGE_STUDY)
T=0.01007(316/31388,GnomAD)
T=0.01182(146/12352,ALFAProject)
T=0.0040(20/5008,1000G)
T=0.0100(45/4480,Estonian)
T=0.0138(53/3854,ALSPAC)
T=0.0135(50/3708,TWINSUK)
T=0.0007(2/2922,KOREAN)
T=0.017(17/998,GoNL)
T=0.017(10/600,NorthernSweden)
T=0.004(2/534,MGP)
T=0.030(9/304,FINRISK)
T=0.07(3/40,GENOME_DK)
A=0.5(2/4,SGDP_PRJ)
T=0.5(2/4,SGDP_PRJ)
High/Med VUS Variants
C=0.000446(112/251276,GnomAD_exome)
C=0.000271(34/125568,TOPMED)
C=0.000388(47/121012,ExAC)
C=0.00015(12/78702,PAGE_STUDY)
C=0.00102(32/31394,GnomAD)
C=0.00031(4/13006,GO-ESP)
C=0.00072(8/11176,ALFAProject)
C=0.0002(1/5008,1000G)
C=0.0029(13/4480,Estonian)
C=0.0010(4/3854,ALSPAC)
C=0.0003(1/3708,TWINSUK)
G=0.109240(27472/251484,GnomAD_exome)
G=0.101156(12702/125568,TOPMED)
G=0.106599(12942/121408,ExAC)
G=0.06797(5349/78702,PAGE_STUDY)
G=0.09949(3120/31360,GnomAD)
G=0.13586(1977/14552,ALFAProject)
G=0.0731(366/5008,1000G)
G=0.1373(615/4480,Estonian)
G=0.1365(526/3854,ALSPAC)
G=0.1429(530/3708,TWINSUK)
G=0.0488(143/2930,KOREAN)
G=0.0448(82/1832,Korea1K)
G=0.135(135/998,GoNL)
G=0.033(20/614,Vietnamese)
G=0.138(83/600,NorthernSweden)
G=0.199(106/534,MGP)
G=0.102(31/304,FINRISK)
G=0.076(23/302,HapMap)
G=0.130(28/216,Qatari)
C=0.49(43/88,SGDP_PRJ)
G=0.15(6/40,GENOME_DK)
C=0.50(7/14,Siberian)
G=0.50(7/14,Siberian)
A=0.033212(8344/251236,GnomAD_exome)
A=0.053369(7701/144298,ALFAProject)
A=0.038258(4804/125568,TOPMED)
A=0.032432(3937/121394,ExAC)
A=0.01389(1093/78702,PAGE_STUDY)
A=0.03825(1200/31372,GnomAD)
A=0.0126(63/5008,1000G)
A=0.0426(191/4480,Estonian)
A=0.0794(306/3854,ALSPAC)
A=0.0690(256/3708,TWINSUK)
A=0.0072(15/2084,HGDP_Stanford)
A=0.0189(23/1214,HapMap)
A=0.0201(22/1094,Daghestan)
A=0.054(54/998,GoNL)
A=0.018(11/626,Chileans)
A=0.075(45/600,NorthernSweden)
A=0.032(17/534,MGP)
A=0.033(10/304,FINRISK)
A=0.005(1/216,Qatari)
A=0.03(1/40,GENOME_DK)
G=0.50(6/12,SGDP_PRJ)
A=0.50(6/12,SGDP_PRJ)
G=0.5(1/2,Siberian)
A=0.5(1/2,Siberian)
T=0.010239(2575/251490,GnomAD_exome)
T=0.009573(1202/125568,TOPMED)
T=0.010090(1225/121410,ExAC)
T=0.00410(323/78702,PAGE_STUDY)
T=0.01007(316/31388,GnomAD)
T=0.01182(146/12352,ALFAProject)
T=0.0040(20/5008,1000G)
T=0.0100(45/4480,Estonian)
T=0.0138(53/3854,ALSPAC)
T=0.0135(50/3708,TWINSUK)
T=0.0007(2/2922,KOREAN)
T=0.017(17/998,GoNL)
T=0.017(10/600,NorthernSweden)
T=0.004(2/534,MGP)
T=0.030(9/304,FINRISK)
T=0.07(3/40,GENOME_DK)
A=0.5(2/4,SGDP_PRJ)
T=0.5(2/4,SGDP_PRJ)
C=0.000690(173/250788,GnomAD_exome)
C=0.000788(99/125568,TOPMED)
C=0.000678(81/119478,ExAC)
C=0.00038(30/78644,PAGE_STUDY)
C=0.00064(20/31392,GnomAD)
C=0.00054(7/13004,GO-ESP)
C=0.00098(11/11176,ALFAProject)
C=0.0010(4/3854,ALSPAC)
C=0.0011(4/3708,TWINSUK)
C=0.001(1/998,GoNL)