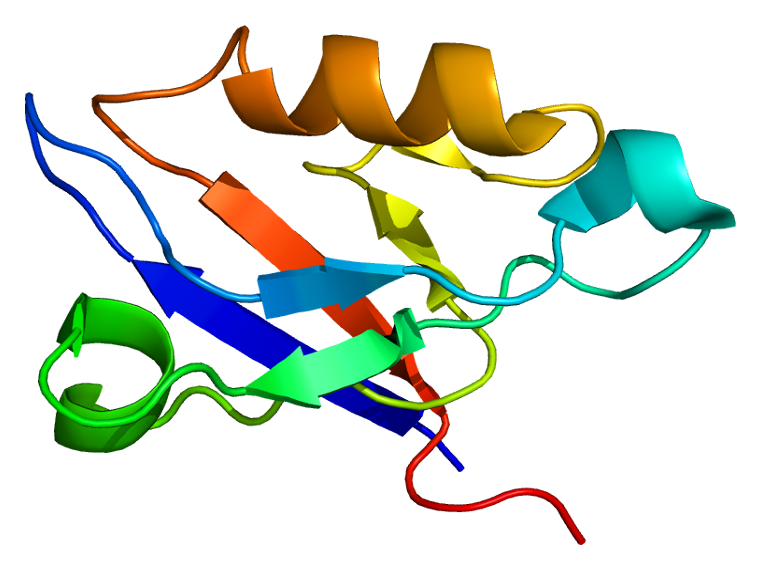
This gene encodes a PDZ domain-containing protein. PDZ motifs are modular protein-protein interaction domains consisting of 80-120 amino acid residues. PDZ domain-containing proteins interact with each other in cytoskeletal assembly or with other proteins involved in targeting and clustering of membrane proteins. The protein encoded by this gene interacts with alpha-actinin-2 through its N-terminal PDZ domain and with protein kinase C via its C-terminal LIM domains. The LIM domain is a cysteine-rich motif defined by 50-60 amino acids containing two zinc-binding modules. This protein also interacts with all three members of the myozenin family. Mutations in this gene have been associated with myofibrillar myopathy and dilated cardiomyopathy. Alternatively spliced transcript variants encoding different isoforms have been identified; all isoforms have N-terminal PDZ domains while only longer isoforms (1, 2 and 5) have C-terminal LIM domains. [provided by RefSeq, Jan 2010]
- Heart development
- Actin cytoskeleton organization
- Sarcomere organization
- Muscle structure development
- Cardiomyopathy, dilated, 1c, with or without left ventricular noncompaction
- Myopathy, myofibrillar, 4
- Myofibrillar myopathy
- Familial isolated arrhythmogenic ventricular dysplasia, right dominant form
- Familial isolated arrhythmogenic ventricular dysplasia, biventricular form
- General Cardiomyopathy
- Hypertrophic Cardiomyopathy
- Dilated Cardiomyopathy
Based on Ayass Bioscience, LLC Data Analysis
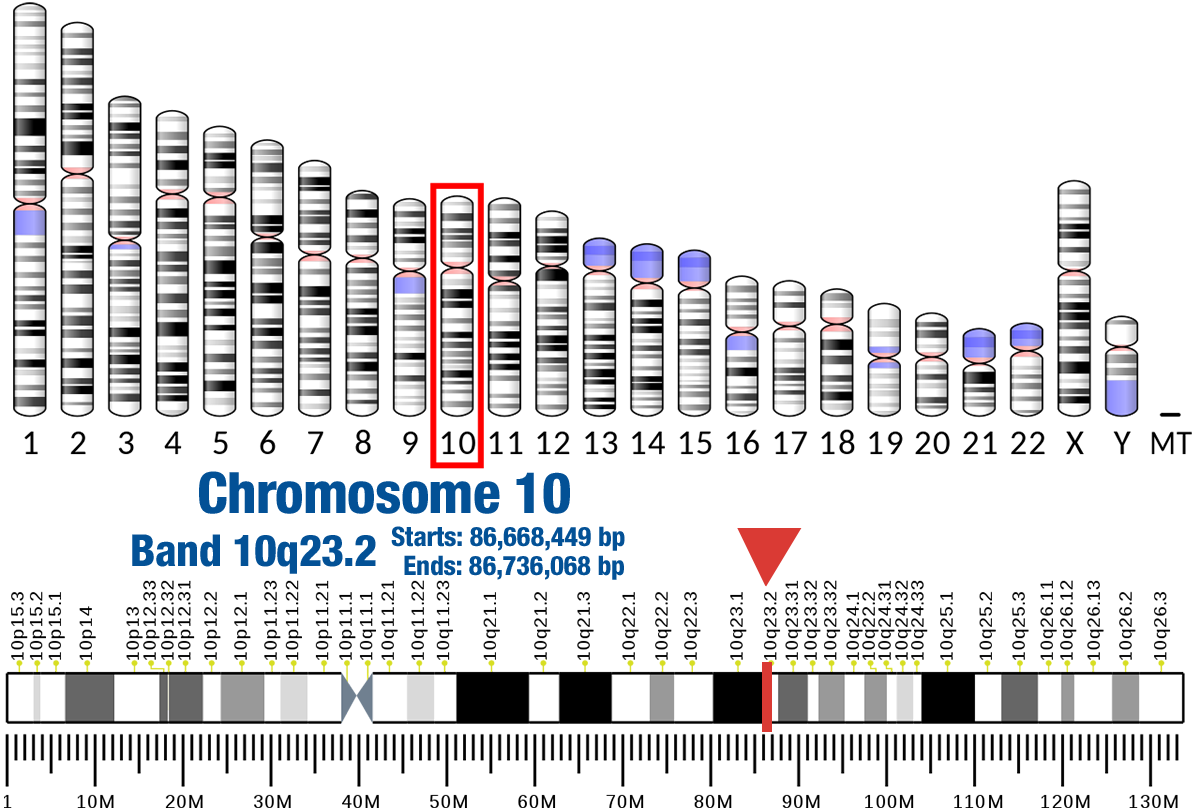
Gene Location
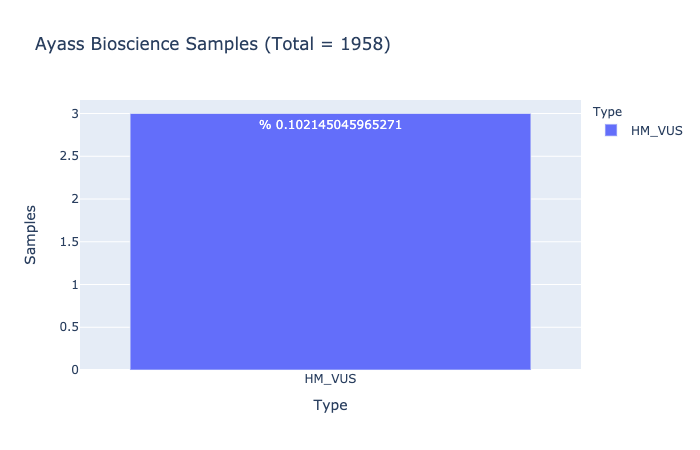
HM-VUS Prevalence
% 0.102145045965271
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
High/Med VUS Variants
T=0.000076(19/251488,GnomAD_exome)
T=0.000104(13/125568,TOPMED)
T=0.000091(11/121404,ExAC)
T=0.00009(9/94990,ALFAProject)
T=0.00006(5/78702,PAGE_STUDY)
T=0.00006(2/31400,GnomAD)
T=0.00023(3/13006,GO-ESP)
C=0.5(1/2,SGDP_PRJ)
T=0.5(1/2,SGDP_PRJ)
C=0.005026(1253/249284,GnomAD_exome)
C=0.003257(409/125568,TOPMED)
C=0.005485(661/120504,ExAC)
C=0.00268(84/31340,GnomAD)
C=0.00320(41/12818,GO-ESP)
C=0.00814(91/11176,ALFAProject)
C=0.0034(17/5008,1000G)
C=0.0007(3/4480,Estonian)
C=0.0042(16/3854,ALSPAC)
C=0.0030(11/3708,TWINSUK)
C=0.009(9/998,GoNL)
C=0.002(1/534,MGP)
C=0.005(1/216,Qatari)
T=0.5(2/4,SGDP_PRJ)
C=0.5(2/4,SGDP_PRJ)