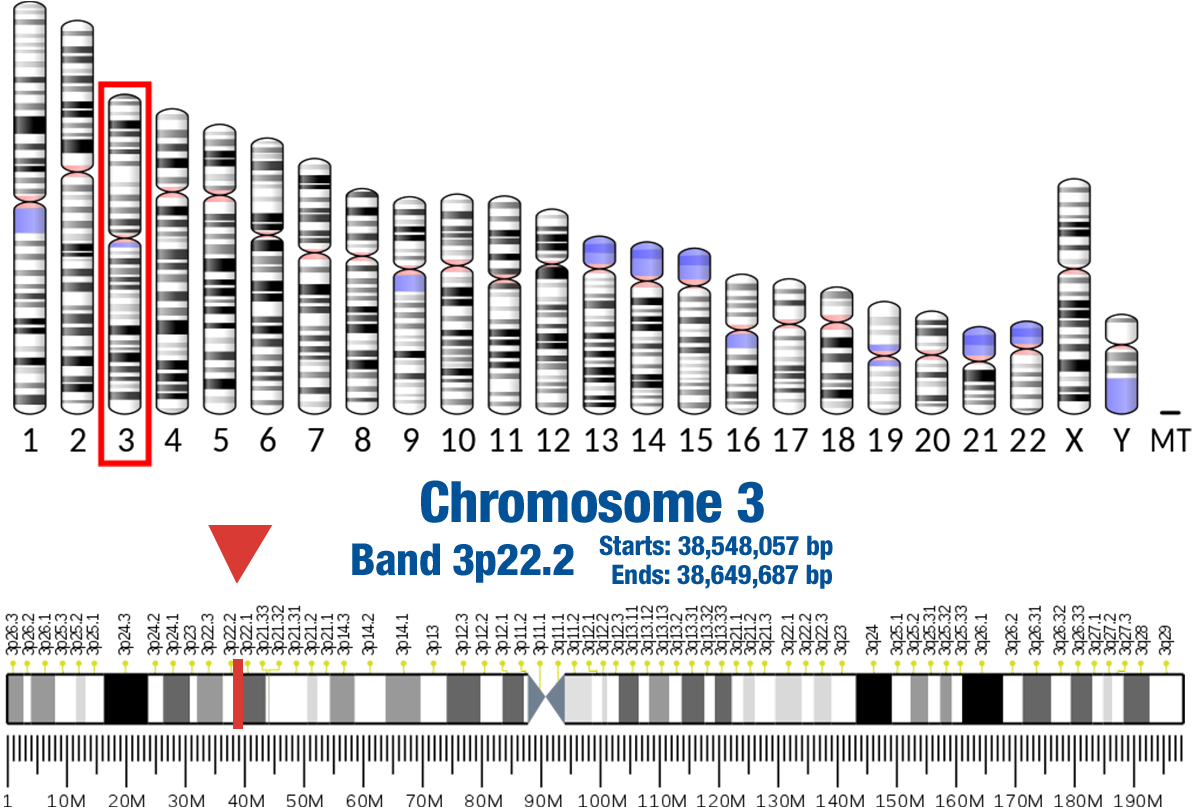
Gene Location
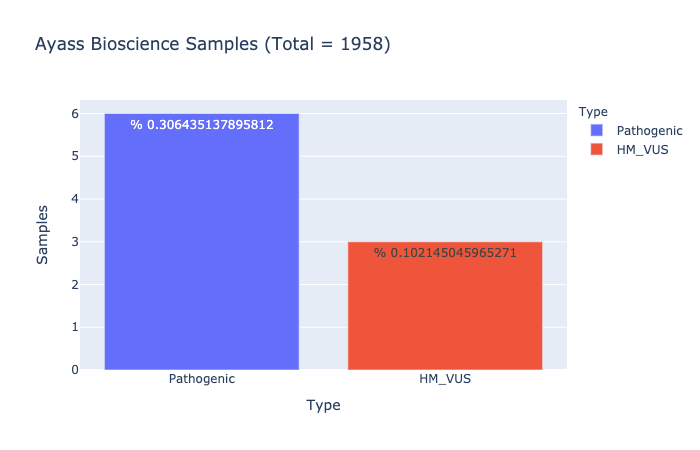
Pathogenic Prevalence
% 0.306435137895812
Ratio of samples with at least 1 pathogenic variant (Computed from Ayass Bioscience Samples)
HM-VUS Prevalence
% 0.102145045965271
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)